Generic Model of Eukaryotic Cell Cycle Control: Biological Details
This page gives further description on the biology behind the generic model of cell cycle control.
Cell cycle control in eukaryotes.
The cell division cycle is the periodic repetition of a sequence of events - including DNA synthesis, mitosis and cytokinesis - that transform a single cell into two daughter cells.
Cell reproduction requires, first and foremost, that the genetic information stored in the cell's DNA be copied and distributed between the two progeny cells. In eukaryotes, DNA copying and distributing are carried out in separate periods of time, called S phase (DNA synthesis) and M phase (mitosis). The temporal gaps between the S and M phases are called G1 and G2. Successful completion of the cell cycle requires that the major events - DNA replication, mitosis and cell division -occur in that specific order. If a cell cycle event cannot be completed (say, the cell's DNA is damaged or its replication is hindered), then the subsequent events of mitosis and cell division must be delayed.
In addition, cell division must proceed in synchrony with the doubling in cytoplasmic mass to achieve a balanced growth and division. If this requirement is not satisfied, then proliferating cells become progressively smaller or larger, which can happen for a limited period of time (as in oogenesis and embryogenesis) but not indefinitely.
Basic components of the cell cycle engine is highly conserved.
Although the complexities of the organisms increase tremendously from simple unicellular organism like budding yeast and fission yeast, to insects like fruit flies, to vertebrates like frogs, and to mammals like humans, the genes and proteins involved in cell cycle regulation, as well as their interactions, are well conserved throughout evolution (Nurse 1990).
The master controller is the cyclin-dependent kinase.
The three major cell cycle events - DNA replication, mitosis and cell division, are controlled by cyclin-dependent kinases (Cdks), which are active only when bound to a cyclin partner.
Yeast cells have only one essential Cdk subunit, referred to as Cdk1, and they use different B-type cyclins in combination with the same Cdk1 subunit to initiate S phase and M phase. The situation in mammalian cells is more complex, as they have other important Cdks (Cdk2, 4 and 6) in addition to Cdk1. In budding yeast, Cdk1 binds with Clb5 (an S-phase cyclin) to form a Cdk1/Clb5 complex (also called S-phase promoting factor or SPF for short) which can phosphorylate proteins at the origin of replication and trigger the onset of DNA replication. By binding with Clb2 (an M-phase cyclin) and forming a Cdk1/Clb2 complex (referred to as M-phase promoting factor, MPF), Cdk phosphorylates histones and initiates mitosis. MPF can substitute for SPF in initiating S phase, but the reverse is not possible.
SPF and MPF not only promote cell cycle transitions but also inhibit them (Dahmann et al. 1995; Botchan 1996; Wuarin and Nurse 1996). For example, once DNA synthesis is initiated by SPF and MPF, the origin of replication loses a necessary component (called the "licensing factor"). Both SPF and MPF inhibit re-loading of the licensing factor, thereby prevent spent origins from being activated a second time until the cell has completed mitosis and destroyed all activities related to SPF and MPF. This dual signalling guarantees the alternation of S and M phases. Likewise, MPF activity promotes entry into mitosis and preparation for cell division, but it inhibits exit from mitosis and completion of cell division (Simanis 2003). Hence, cell division occurs only if the cell has first entered mitosis and then exited.
Mechanisms that regulate Cdk activity.
Cdk/cyclin activities (SPF and MPF) are subjected to multiple levels of regulation. In principle, cells could regulate the availability of the Cdk subunits, but this is uncommon. Most Cdks are present in constant (and excess) abundance throughout the cell cycle. Instead, their activities are regulated in three ways:
- By the availability of cyclin partners. Cyclin abundance is determined by its rate of synthesis and degradation, both of which are regulated during the cell cycle.
- By binding to a stoichiometric inhibitor (referred to as CKI, cyclin-dependent kinase inhibitor). CKI comes and goes, as its synthesis and degradation are also cell cycle regulated.
- By phosphorylation of a specific tyrosine residue in the kinase subunit. The phosphorylation state of Cdk varies during the cell cycle as the activities of the tyrosine kinase (Wee1) and the tyrosine phosphatase (Cdc25) fluctuate.
Interactions of Cdk and its regulators involve many feedback loops.
Interestingly, all the regulators of the Cdk activity are themselves subject to controls by Cdk activities, creating positive and negative feedback loops in the mechanism (see the wiring diagram and the corresponding differential equations for the model). For example, in budding yeast, the transcription factor that stimulates CycB (a cyclin important for indcing mitosis) synthesis is turned on by CycB/Cdk, creating a positive feedback loop; whereas the Cdc20/APC complex (which is partially responsible the proteolytic degradation of CycB) is activated by CycB/Cdk, creating a negative feedback loop. Also see Table 1 below.
Species-specific regulations of the Cdk network.
Table 1 summarizes the components of the regulatory network in the budding yeast, fission yeast and mammalian cells. It also describes their functions and how they interact with the master cell cycle regulator, the Cdk kinases.
 Table 1. Name conversion table for major cell cycle regulators.
Table 1. Name conversion table for major cell cycle regulators.
Table 2 shows the parameter values used in simulations for these organisms. It can be seen that, although the regulation mechanism is quite conserved, there are specific differences among different organisms.
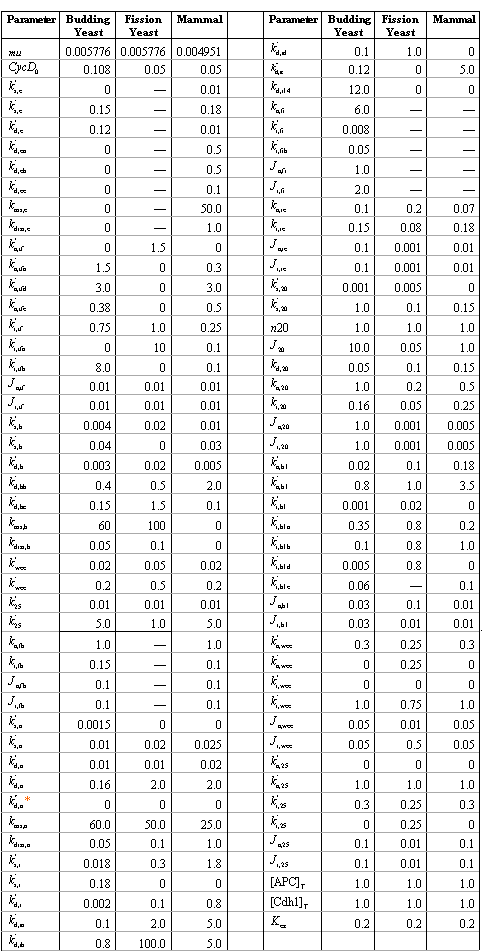
Table 2. Parameters used in the generic model.
Such differences in reaction mechanisms and rate constants reflect the different mutant phenotypes in these organisms. For instance, deletion of Wee1 gene or Cdc25 gene has little affect on their cell cycle progression in budding yeast, the two mutant cells are viable, with sizes close to that of the wild type. In fission yeast, however, deletion of Wee1 gene makes the mutant cell half the size of the wild type; deletion of Cdc25 gene makes the mutant inviable, arrested as a long rod in G2 phase.
This is because, in fission yeast, the mitotic CycB is continously made throughout the cell cycle; whereas in budding yeast, the transcription factor for CycB is activated by MPF, so its peak of synthesis occurs late in G2. In order to prevent premature entering into mitosis, the fission yeast cells, however, rely on the inhibitory kinase Wee1 to keep Cdk1 phosphorylated and inactive. Wee1 is inhibited and Cdc25 activated only when the cell has grown to a critical size for triggering mitosis. As a consequence, deletion of Wee1 causes the cell to enter mitosis prematurely, resulting in small cell size. Deletion of Cdc25, on the other hand, prevents the cell from entering mitosis, resulting in a G2 arrest.
Mathematical modeling with the help of JigCell is needed for analysis of complex networks.
Mathematical modeling, based on biochemical rate equations, provides a rigorous and reliable tool for unraveling the complexities of regulatory networks, such as the cell cycle control mechanism described here.
JigCell was developed with this goal in mind, to help modelers to construct accurate mathematical models from the wiring diagrams, to run batch simulations, and to facilitate the task of finding the proper parameter set by automatically comparing the simulation results with experimental observations, and eventually to search the parameter space in order to derive an optimal parameter set.