Generic Model of Eukaryotic Cell Cycle Control: Mathematical Model
Molecular biology of cell cycle regulation in eukaryotes
The cell division cycle is the sequence of events during which a growing cell replicates all its components and divides them between two daughter cells, so that each daughter contains the information and machinery necessary to repeat the process.
To ensure that the genetic material, the DNA molecules, is dutifully copied and evenly distributed between the two progeny cells, and to ensure size homeostasis, three basic requirements for the successful completion of a cell cycle must be met:
- the DNA-synthesis phase (S phase) and the DNA-distribution phase (mitosis or M phase) must alternate,
- DNA replication checkpoint and DNA damage checkpoint must be operative to arrest the cell cycle before mitosis, until replication is complete and DNA is undamaged,
- and the DNA cycle (S->M->S->M) must coordinate with the growth cycle.
Satisfying these three requirements is not a characteristic of any single component of the cell cycle machinery, but rather an emergent property of the way the components interact. Control of the sequence and timing of cell cycle events is a complex and vital task as described briefly below.
The genes and proteins involved in cell cycle regulation, as well as their interactions, are well conserved throughout evolution from yeast to frogs to mammals (Nurse 1990). These three major cell cycle events - DNA synthesis, mitosis and cell division - are controlled by a family of protein kinases called cyclin-dependent kinases (Cdks for short). The name for these kinases comes from the fact that they are active only when bound to a regulatory subunit called cyclin. There are various cyclins in the cells. Cyclin binding determines the substrate specificity of the Cdk enzyme. Cdk, when paired with suitable cyclin partner, phosphorylates particular target proteins and thereby triggers specific cell cycle event.
Cdk activities are regulated throughout the cell cycle in many ways:
- the availability of cyclin partners can be controlled by synthesis and degradation,
- the activity of the Cdk/cyclin dimer can be further modified by phosphorylation of the Cdk kinase subunit,
- and the fomation of an inactive Cdk/cyclin/CKI trimer molecule with a cyclin-dependent kinase inhibitor (CKI).
It is interesting that the enzymes that regulate the Cdk activity are all regulated in turn by Cdk activity. Some are activated by Cdk activity, some are inhibited by it. Thus there are many positive and negative feedback loops in the regulatory network.
The general mechanism of how an eukaryotic cell controls its Cdk activites is shown in Fig. 1 below.
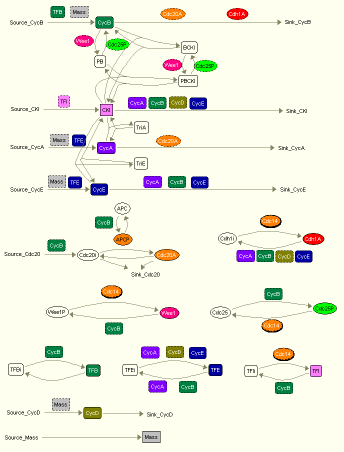
Fig. 1. Wiring diagram of a generic model for eukaryotic cycle cell conrol, created by JDesigner.
Although the scheme of cell cycle control is quite conserved, there are species-specific differences, giving rise to specifc behavior of the control network when certain genes are mutated. Table 1 summarizes the components of the regulatory network in the budding yeast, fission yeast and mammalian cells. It also describes their functions and how they interact with the master cell cycle regulator, the Cdk kinases.

Table 1. Name conversion table for major cell cycle regulators.
Table 2 summarizes the parameter value used in the simulations of the generic model for the budding yeast, fission yeast and mammalian cells. [Click on Table 2 for a full list]

Table 2. Parameters for the generic model.