Generic Model of Eukaryotic Cell Cycle Control: Validation
The JigCell Comparator is a tool for performing automatic, quantitative comparison between experimental data and model simulations. A modeler may explore the parameter space manually using the Comparator and one of its accessories, the Report Generator. By automating the comparison process, the Comparator takes a large part of the drudgery out of the parameter-twiddling stage, the most time-consuming stage of model exploration. It also provides the ground work for the next stage: automatic parameter estimation.
Before running the Comparator, the user first runs facilitator.exe (located in JigCell/facilitator directory) which enables the simulator to talk with the Run Manager, then runs xppserver.bat (in the JigCell directory), which opens the simulator. Then comparator.bat (in the JigCell directory) starts the Comparator.
The Comparator uses a spreadsheet interface. There are three main tabs, labeled as Experiment, Transform and Objectives, respectively.
On the Experiment spreadsheet (see Fig. 14), the user starts from the Edit menu to insert rows for data entry. Then he enters data for the first row (the first experiment), in the following order: the name of the experiment (in column #1), the experimental data (in column #2), and its value type (in column #3) which corresponds to the algorithm used for comparing simulations to experiments. When the user right-clicks in the cell for Experimental Value and selects the option Edit, a window will pop up, where he can enter the relevant experimental data for comparison. Once the user completes the first row, he can right click on that row, select Fill Down to enter and edit data for similar experiments. The data are stored in the file model.dat.
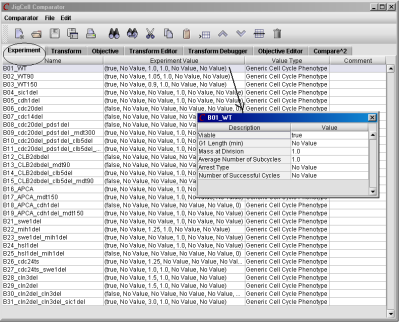
Fig. 14. The Experiment spreadsheet describes the experimental data.
On the Transform spreadsheet (see Fig. 15), each row associates an experimental observation with a name of a transform describing how to simulate such an experiment (by assigning a name of the run in the run file) and how to how to extract from the time courses information that can be compared with experimental observations. The transforms are stored in the file model.mss.
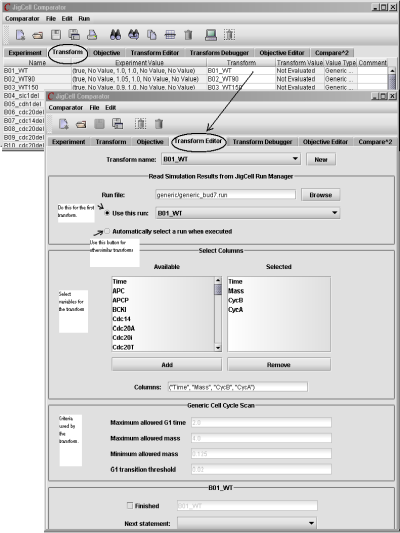
Fig. 15. The Transform spreadsheet and the Transform Editor.
On the Objectives spreadsheet (see Fig. 16), each row describes how to compute a numerical value representing the goodness-of-fit of the simulation to data, and specifies a threshold number for acceptance. Unacceptable fits are highlighted in red.
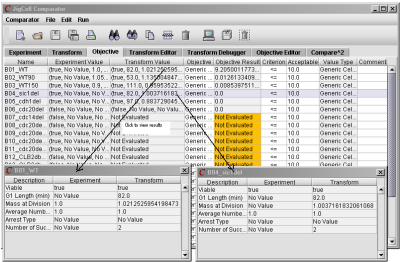
Fig. 16. The Objectives spreadsheet and a partial result from the Comparator.
The Report Generator is a utility program in the Comparator which creates a PDF file for a set of simulations specified by the user. Fig. 17 shows how to generate such a report after the comparator has finished its task.
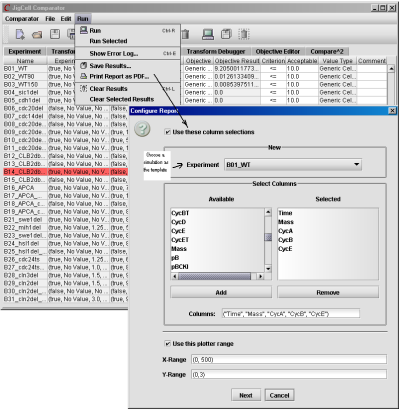
Fig. 17. An illustration of how to use the Report Generator to create a PDF file.
Click on the links to see a screen shot of the Comparator results for budding yeast, fission yeast, and mammalian cells.
Comparator indicates unacceptable fits in red, so the modeler can see at a glance where the model (equations or parameter values) or the transform is having problems. In the case for budding yeast, the error in predicting the viability of the the mutant B14_Clb2dbdel_clb5_del is becasue the model is too simple, it fails to take into account of the effect of the DNA unreplication checkpoint. In fission yeast, the error in mutant P07_cig2del is due to a transform problem, whereas the error in mutant P17_pUC1_cig2del is due to problems in parameter values. In all these cases, the Comparator detects the errors quickly, and points out areas that need further modification.
Exploring the parameter space in searching for an optimal parameter set is the most common and tedious work in mathematical modeling. With the help of JigCell, the modeler can now quickly see how a change of the numerical values of some parameters propagates through the entire model and affects all the output data. This greatly facilitates the progress of model development, and sets the stage for the development of the ultimate tool for automatic parameter estimation.